SL Screens
Investigating the use of screens in Super Learner ensembles
Drew King, Brian D Williamson, Ying Huang
Investigating the use of screens in Super Learner ensembles
Drew King, Brian D Williamson, Ying Huang
Abstract
Clinical research trials often generate large datasets with many variables, which are analyzed to identify potential relationships with clinical outcomes. Machine learning techniques are increasingly employed for such analyses, yet little is known about the accuracy of these algorithms in different settings. This research aims to establish guidelines for combining variable selection algorithms to assess accuracy under various conditions. Our findings indicate that using an ensemble of algorithms with the Lasso variable selection tool can produce inconsistent accuracy with certain types of datasets. This research was published to The New England Journal of Statistics in Data Science on May 7th 2025, additionally the paper is available with open access on arXiv.org. The experiment can be reproduced using simulation code on GitHub written by Doctor Brian Williamson.
Note: All code is open-source under the MIT licene.
Introduction
Clinical research often involves large-scale studies, such as those on vaccines or cancer early detection, but rare diseases can result in smaller sub-samples. This imbalance complicates variable selection because many existing tools are not optimized for datasets where some outcomes or categories are uncommon.
Despite these challenges, machine learning has become a mainstay in biomedical research because it can handle a large number of variables and detect complex relationships between variables and clinical outcomes. However, performance varies significantly across different algorithms, suggesting that no single approach is universally optimal. One proposed solution is to adopt an ensemble method—combining outputs from multiple algorithms—to leverage their individual strengths. At present, there are no widely accepted guidelines on how best to combine these procedures, creating a need for clear recommendations. Our goal is to develop such guidelines, potentially improving research in domains like vaccine efficacy and cancer detection.
Contributions
Brian Williamsom, PhD
- Designed the experiment and wrote the simulation code in R.
- Mentored Drew in developing analysis code.
- Authored the research paper and abstract.
Drew King, BSc
- Collaborated on deploying the experiment to a Linux server cluster using bash.
- Created R scripts (with tidyr, dplyr, and ggplot2) to aggregate and visualize results.
- Designed a poster to present the research to MESA (an organization for minority students in STEM).
Ying Huang, PhD
- Provided guidance on experimental design and data analysis.
- Offered feedback and direction on the research paper.
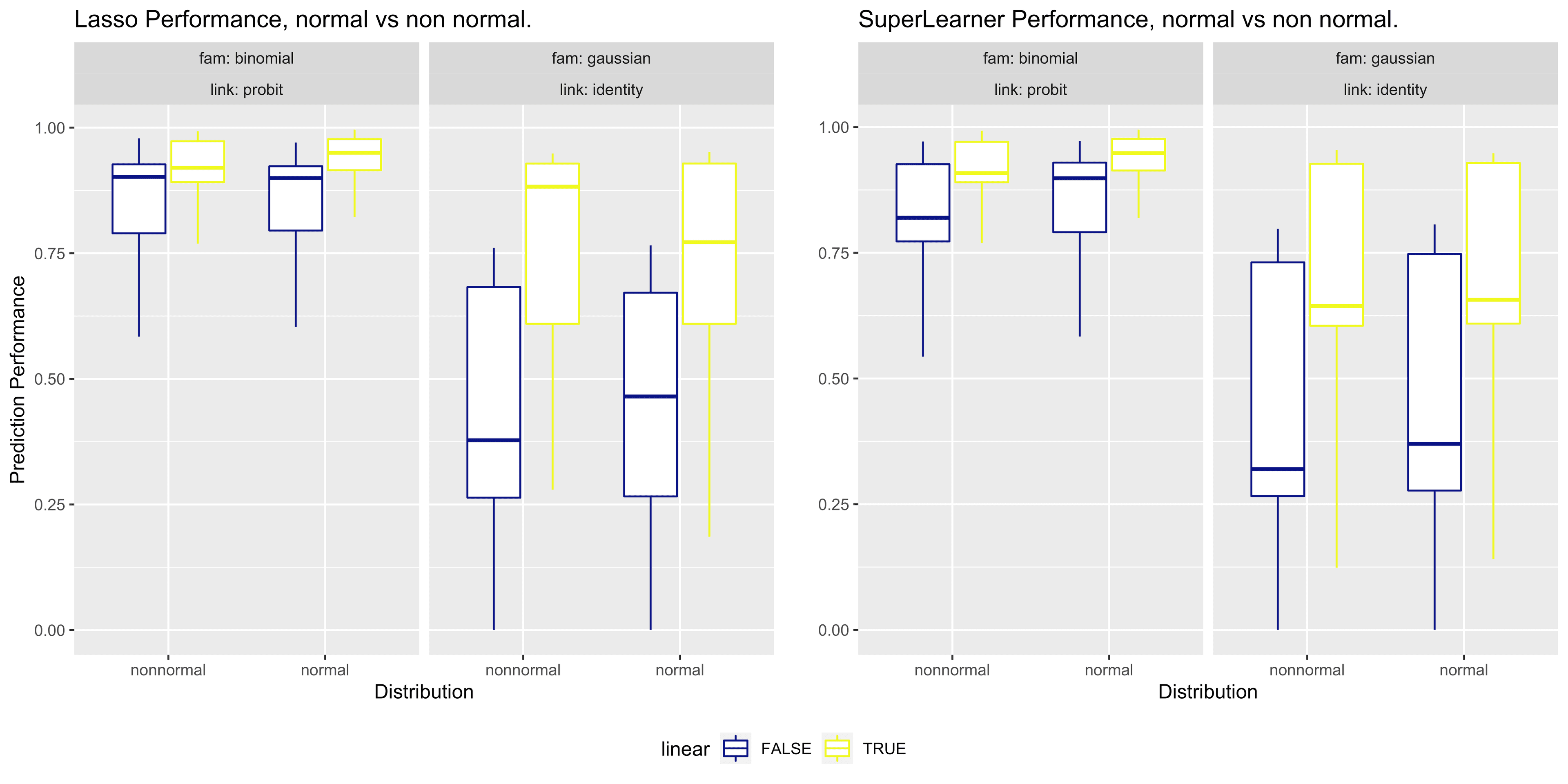
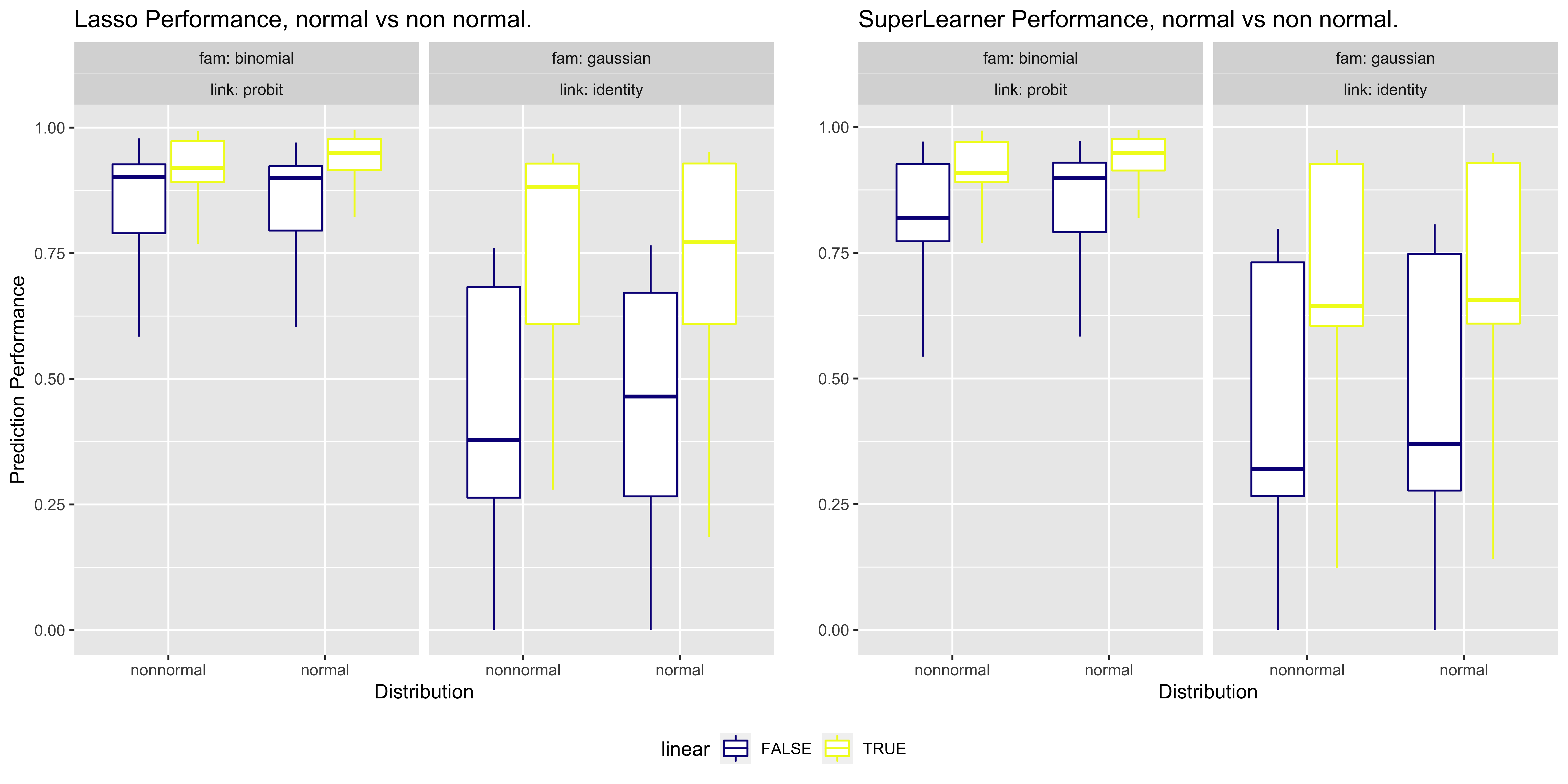
Results
Our preliminary analysis shows that Lasso + Super Learner (SL) is not always a beneficial combination, which is a surprising outcome. In particular:
- SL with appropriate screening mechanisms (or with Lasso and screens) tends to be robust for most datasets.
- Lasso alone can be unreliable in many instances.
- Lasso + SL without any screening is especially inconsistent in certain cases.
Next Steps
We plan to train researchers on applying these ensemble guidelines and continue developing our methods. Future work includes:
- Investigating why Lasso alone often underperforms for variable selection.
- Testing other algorithms that are optimized for biomedical data.
- Rewriting the experiment in faster languages (e.g., C or Python) for improved performance.
- Designing interactive data visualizations using D3.js to facilitate more intuitive result exploration.
Our results are fully reproducible with standard scientific computing tools in R, allowing others to validate and extend our findings.